Retroelement and Retrovirus Universal Classification
- Pat Heslop-Harrison
Retroelements and Related
Viruses - A Classification
Here, we give a classification
of retroelements and related viruses (presented in C Hansen and JS
Heslop-Harrison, 2004. Sequences and phylogenies of plant pararetroviruses,
viruses and transposable elements. Advances in Botanical Research 41:
165-193: see this link for text including colour figures and supplementary information; and based on work by Hull,
1999a, 2001; and ICTV International Committee for the Taxonomy of Viruses)
2003.
Nuclear DNA elements that
include the gene for reverse transcriptase, RT, (‘retroelements’,
also known as class I transposable elements) can be integrated into a unified
classification for reverse transcribing elements that includes viruses and transposable
elements (Hull
1999). The elements can be divided into retroviruses, pararetroviruses and
the abundant group of nuclear sequences, the retrotransposons, including the
long terminal repeat (LTR) retrotransposons, non-LTR retroposons and group II
mitochondrial introns (Hull, 1999a, 2001). Another important enzyme,
telomerase, which adds the telomere sequences to most eukaryotic chromosomes,
also incorporates a RT function
The diagram below shows the
relationship of all these elements. At the bottom of the page, an alignment
of the reverse transcriptase domain of representatives of most of these
suborders shows how all the amino acid sequences has conserved active sites..
Abbreviations: CaMV, Cauliflower mosaic caulimovirus; BSV, Banana streak badnavirus; SbCMV, Soybean chlorotic mottle soymovirus; PVCV, Petunia vein clearing petuvirus; CsVMV, Cassava vein mosaic cavemovirus; RTBV, Rice tungro bacilliform tungrovirus; TVCV, Tobacco vein clearing cavemovirus. The taxonomic endings follow
the ICTV nomenclature: order - virales;
suborder (after Hull)
- ineae; family - viridae; subfamily (not shown) - virinae; genus - virus. Mis-spelling: retorelements, retrorelements, retroelments, retrelements, retroelemnts (Link to Universal protocols for isolation of retroelement fragment from genomic DNA by PCR)
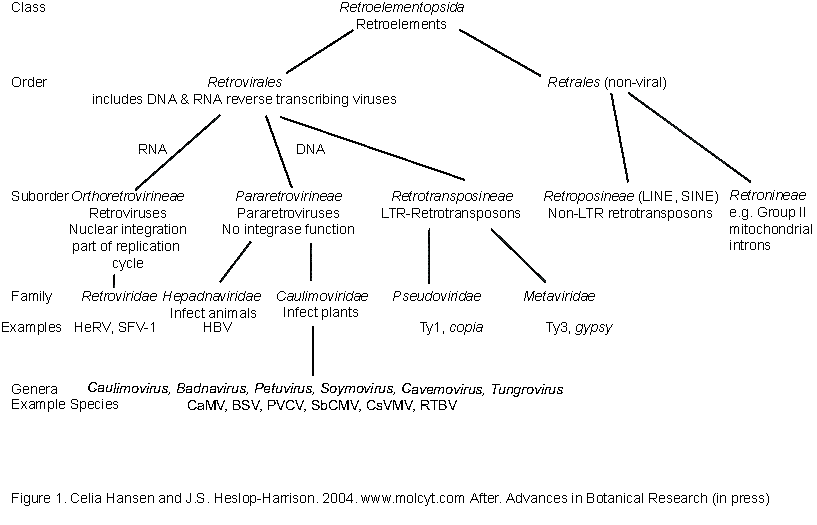
After Figure 1. Celia Hansen and J.S.
Heslop-Harrison. 2004. www.molcyt.com After. Advances in Botanical Research
41: 165-193.
(Diagram above); for Indexing
programmes, lines are as follows in text:
CLASS: Retroelementopsida (Retroelements)
ORDERS: Retrovirales (includes DNA and RNA reverse
transcribing viruses) AND Retrales (non-viral)
ORDER
Retrales
Suborder Retroposineae (LINE, SINE) Non-LTR
retrotransposons
Suborder Retronineae (e.g. Group II mitochondrial
introns)
ORDER
Retrovirales
Suborder
Orthoretrovirineae (RNA; Retroviruses Nuclear integration part of replication
cycle Example family Retroviridae Examples HeRV, SFV-1)
Suborder
Pararetrovirineae (DNA; Pararetroviruses, No integrase function)
Family:
Hepadnaviridae (infect animals, e.g. HBV)
Family:
Caulimoviridae (infect plants)
(Caulimoviridae
genera are Caulimovirus, Badnavirus, Petuvirus, Soymovirus, Cavemovirus,
Tungrovirus, an example virus in each genus is CaMV, BSV, PVCV, SbCMV, CsVMV,
RTBV)
Suborder Retrotransposineae
(DNALTR-Retrotransposons)
Family:
Pseudoviridae (e.g. Ty1, copia)
Family:
Metaviridae (e.g. Ty3, gypsy)
An
alignment of amino acid sequences from a range of different nuclear elements
containing reverse transcriptase. Conserved residues are marked in red, and
the domains relate to known and postulated functional regions of the
sequence. Sorry – you will need to drag out your browser window to show
the full lines of the alignment and then use the scroll bar at the bottom of the screen.
< domain 3 > < domain 4 > < domain 5 > < domain 6 > < domain 7 > Eap123 MDIEKCYDSVNREKLSTFLKTTK103FYKQTKGIPQGLCVSSILSSFYY·······NVNLLMRLTDDYLLITTQ··FIEKLINVSRENGFKFNMK KLQTSF······ NIVQDYCDWIGISIDMKTLAL BLIN LDLARAFDSVSWPFLFEVLRCHG27PAIWHRRGLHQGDPVSPQLLVLAV········IPAISLYADDVILLCHP··AVKEILQLFGRASGLHVNFQKSAAAL······ IVDFPLTYLGIPLKLRRPTAGQLQ Ty1 LDISSAYLYADIKEELYIRPPPH·NDKLIRLKKSLYGLKQSGANWYETI········QVTICLFVDDMVLFSKN LNSNKRIIEKLKMQYDTKIINLGESDEEIQYDILGLEIKYQRGKYMKLGMENS Copia MDVKTAFLNGTLKEEIYMRLPQG·· NVCKLNKAIYGLKQAARCWFEVF··········YVLLYVDDVVIATGD MTRMNNFKRYLME KFRMTDLNEIKHFIGIRIEMQEDKIYLSQSAYVKKILSKFNM BARE-1 MDVKAAFLNGLLKEELYMMQPEG··· ACKLQGSIYGLVQASRSWNKRF·········AFLILYVDDILLIGNG VEFLENIKDYLNK SFSMKDLGEAAYILGIKIYRDRSRVIGLSQSTYLDKVLKRFK Gypsy LDLKSGYHQIY··EKTSFSV·· FCRLPFGLRNASSIFQRALDDVLREQIGKICYVYVDDVIIFSEN··HIDTVLKCLIDANMRVSQE KTRFFK EYLGFIVSKDGTKS····EPDCVYKVRSFLG BAGY-1 MDLRLGYHQIK··PKKAFVT·· YTVMSFGLTNAPATFSRLMNSIFMEYLDKFVVVYLDDILIYSMN··HLRLVLMKLREHRLYAKFS KCEFWY HKVTYLGHVISGKGIAV····QPESVKQVRSFLG Cyclops LDGYSGYNQIA··*KTAFTC·· YRKMSFGLCNAPTTFQRCVQAIFADLNEKTMEVFMDDFSVFGVS··NLKTVLERCVKTNLVLNW* KCHFMV TEGIVLGHKVSSRGLEV····PPVNVKGIRSFLG Athila LDGYSGFFQIP··EKTTFTC·· YKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVYGPS··NLGRVLTRCEETNLVLNWE KCHFMV KEGIVLDHKISEKGIEV····PPKTVKDIRSFLG CaMV FDCKSGFWQVL··PLTAFTC·· WNVVPFGLKQAPSIFQRHMDE AFRVFRKFCCVYVDDILVFSNN··HVAMILQKCNQHGIILSKK KAQLFK KKINFLGLEIDEGTHKPQGHILEHINK tprv FDCKSGFYHLK··KLTAFTV·· WNVLPFGYKNAPGRYQHFMDN YFNQLENCIIYIDDILLYSRT··LLEKFIHIVEISGISLSKK KAEVMK NQIEFLGIQIDKNGIKMQTHVVQKI BSV FDLKSGFHQVA··PWTAFWA·· WLVMPFGLKNAPAIFQRKMDN CFRGTEDFIAVYIDDILVFSET··HLKKFMTICEKNGLVLSPT KMKIGT RQIDFLGATIGNSKIKLQPHII HBV LDVSAAFYHIP58FGRKLHLYSHPIILGFRKIPMGVGLSPFL LAQFTS·· RRAFPHCVAFSYMDDVVLGAKS··LFTSITNFLLSLGIHLNPN KTKRWG YSLNFMGYVIGSWGTLPQEHI HERV-K IDLKDCFFTIP··EKFAFTI·· EPATRFQWKVLPQGMLNSPTICQTFV··VREKFSDCYIIHYIDDILCAAET··CYTFLQAEVANAGLAIASD KIQTST PFHYLGMQI SFV LDLSNGFWAHS··WLTAFTWLGQQYCW TRLPQGFLNSPALFTADV VDLLKEVPNVQVYVDIYISHDDP··LEKVFSLLLNAGYVVSLK KSEIAQ HEVEFLGFNITKEG
|