|
|
Research Projects of Dr Trude Schwarzacher |
|||||||||||
|
with ·
Detailed
protocols ·
Publication
lists · Past and present lab members · Collaborations Completed Projects |
Genome organization and epigenetic mechanisms in
polyploid plant species Most estimates suggest that 50% of plant species are
recognisable and evolutionary recent polyploids where diploid ancestors can be
identified. My lab is
studying the interaction of genomes, chromosomes and DNA sequences in
polyploids and hybrids in several plant groups including cereals, petunia,
banana and others. My work concentrates on repetitive DNA sequences, both
tandemly repeated satellite sequences and dispersed transposable elements,
their diversity and evolution, as well as their epigenetics as evidenced by
chromatin organisation and methylation. We
speculated that it is the biological significance of epigenetic phenomena that
have lead to the successful evolutionary history of polyploidy and hybrid
plant species. In a recent European
collaboration ((PARADIGM) we have studied
genomic integration of
pararetroviruses that are a recently discovered
repetitive sequences class in plants and in petunia and tomato form a
significant part of the genome. They possibly show a much wider distribution
in plant genomes, but are often degenerate and rearranged. We have recent
evidence for pararetrovirus elements in sugar beet and other angiosperms and
are in the process of sequencing full elements. Pararetroviruses are closely
related to pseudoviridae retroelements and are often found in their physical
proximity. Pararetrovirus activation is linked to disease outbreaks and
patterns of DNA and chromatin methylation and possibly epigenetic silencing
mechanisms. Activation of integrated, but dormant pararetroviruses can be
caused by the introduction of wide hybridization and can lead to disease
outbreaks through tissue culture or environmental stresses as has been shown
for banana. In wheat and related species, we have found differences in cytosine
methylation patterns at symmetrical and asymmetrical sites in the tandemly
repeated 120bp repeat DNA family in diploid and polyploid genomes and we are
currently investigating whether other repetitive DNA families show similar
changes. On the chromosomal level, diploid species of rye and wheat show
uniform methylation patterns while allopolyploids have more unevenly
distributed methylation indicating de novo methylation and
demethylation mechanisms when genomes are combined in polyploid and hybrid
species. In wheat breeding lines, incorporating alien chromatin segments
important to understand such epigenetic changes both at the DNA sequence
level and the global chromatin level to predict the successful Our studies in banana have looked at retroelements and
tandemly repeated DNA sequences, and their genomic flanking sequences were
studied showing differential expression, DNA and histone methylation. The close chromosomal proximity and insertion within
each other postulates a possible link in the evolution of
LTR-retrotransposons, tandem repeats and 5S rRNA genes, but also a role of
retrotransposon sequences in gene regulation. This is supported by variable
DNA methylation patterns and the presence of repeat transcripts.
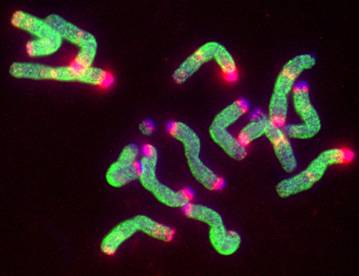
Root tip metaphase of rye, Secale cereal ‘Petkus Sping’ (2n=14) after
immunostaining with anti-methyl cytosine antibodies (green fluorescence)and
fluorescent in situ hybridization
with the 120bp repeat DNA probe (red fluorescence). The 120bp repeat family
is located in the large subtelomeric heterochromatic
blocks that lack DNA methylation while the euchromatin and small intercalary 120bp repeat sites are
uniformly methylated. Schwarzacher and Contento have shown that this uniform methylation pattern gets
disturbed in hybrid and polyploid triticale.
Page updated 24 November 2008 I
have used molecular cytogenetic methods to analyse chromosome behaviour
during early events of meiosis in Triticeae cereals. I have found that
homologous chromosomes first associate at the interphase before meiotic
prophase and have postulated a three stage model of homologous chromosome
pairing in Triticeae cereals (with relatively large genome size) that
involves cognition, alignment and then synapsis. The differential
condensation of pachytene chromosomes and the non-random distribution of
chiasmata, that I have shown in cereal chromosomes, have major implications
for genome organisation, sequence distribution and amplification, chromosome
behaviour and possibly gene expression Evolution
of terminal regions of chromosomes I have studied the long range organisation and possible modes of evolution of repeated sequence families at the telomeres of rye chromosomes, and located microsatellite motifs on metaphase chromosomes in situ. Various motifs show a rich, chromosome-specific and conserved diagnostic banding pattern in rye and wheat. We have found microsatellite repeats occur mainly in the chromatin loops formed at meiotic prophase, in contrast to the subtelomeric satellite sequence families that are closely associated with the synaptonemal complex. Using flow cytometry and sorting of chromosomes, I have found that plant genomes show a remarkable uniformity of AT:GC ratios of all chromosomes, indicative of strong homogenisation events that are different to that found in mammalian genomes. I located the telomeric repeat (TTTAGGG)n on cereal metaphase chromosomes, and showed that different chromosomes carry different amounts of telomeric repeats at their physical ends; I was able to describe unambiguously for the first time the dispersed distribution of retrotransposon-like elements in plants. In situ
hybridisation using total genomic DNA as a probe, developed by me, has enabled
us to identify and localise chromosomes and chromosome segments in cereal
plants that include DNA from different species. The technique, now applied
world-wide by many research labs, has proven to be valuable for studying the
architecture of the nucleus and for following alien gene transfer. We have
shown that the nucleus is non-randomly organised, that individual chromosomes
occupy tightly defined domains at interphase and that the parental genomes in
interspecific and intergeneric hybrids remain separated throughout the cell
cycle. We speculate that there are mechanisms for organising the interphase
nucleus and that this organisation affects chromosome behaviour during gene
expression, chromosome elimination, and during somatic and meiotic division. I have isolated, cloned and sequenced
the meiotic recombination specific gene DMC1
(disrupted meiotic c-DNA, homolgous to RAD51) in barley (in collaboration
with Victor Klimyuk and Jonathan Jones, Sainsbury Laboratory, John Innes Centre).
We used sequence homology from the Arabidopsis gene and primers to the
conserved region to isolate a 11kb lambda genomic clone and have sequenced
most of the gene including the promoter. We have transformed barley to study
the activity and specificity of the DMC1
promoter. Foci with antibodies to Rad51 localise near telomeres of rye
chromosomes indicating that early recombination events are unevenly
distributed along chromosomes and reflect chiasma distribution. I
|
|||||||||||
![[The University of Leicester]](http://www.le.ac.uk/corporateid/department/000066/unilogo.gif)