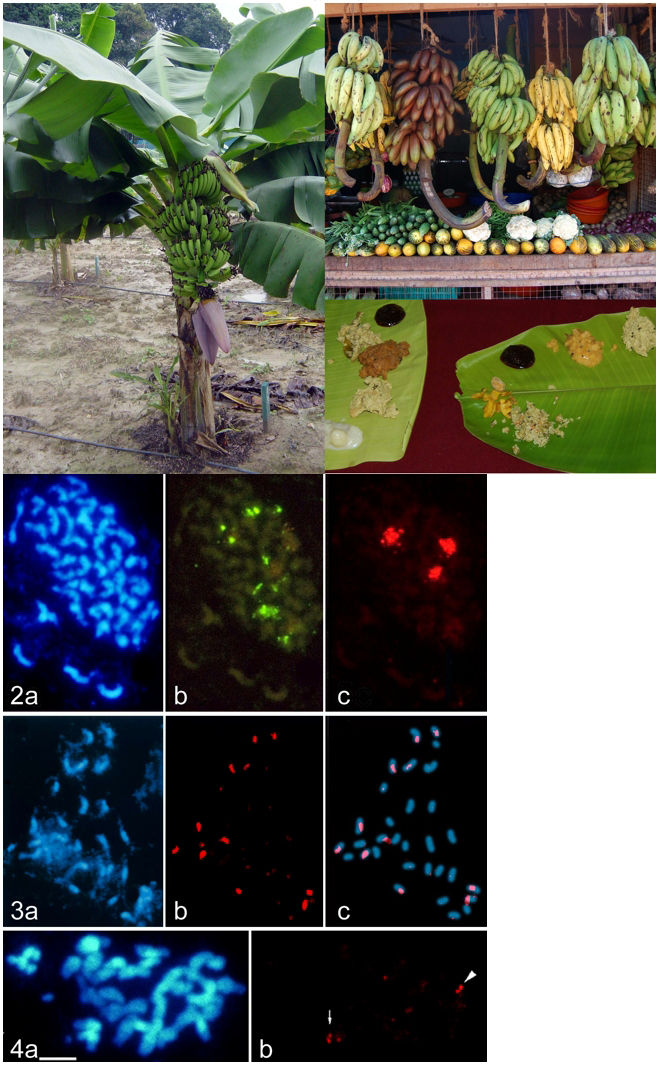
Dessert bananas and the cooking bananas or plantains are among the oldest crops in the world. Most crops were domesticated through a long pathway of selection and crossing, but for banana virtually all the two thousand varieties which are grown throughout the tropics were collected as spontaneous mutants in the wild with the extraordinary property of having large fleshy fruit without any seeds. Banana cytogenetics is also unusual: the varieties that are eaten are mostly triploid hybrids between different species. The wild progenitors of the domesticated banana are Musa acuminata (where the haploid genome is designated as ‘A’) and Musa balbisiana (the B genome). Banana varieties that are hybrids with AAB and ABB genome constitutions are a staple food for a billion people in Asia and Africa and have 2n=3x=33 chromosomes (Figure 1). About 15% of the world’s banana production is for the export trade, and is based on a single variety, ‘Cavendish’. This sweet banana has the genome constitution AAA. Traditionally, bananas are propagated by side suckers to the main stem (figure 1), but in commercial plantations, most plants are now multiplied through tissue culture to ensure disease-free planting material. Abnormal plants may arise; those with gross cytological changes have poor viability and are discarded, but some of the somaclonal mutant variants become successful cultivars such as ‘Dwarf Cavendish’ or ‘Williams’. The different cell layers of the plant meristem are maintained through vegetative propagation, meaning that some desirable colour mutants are actually chimeras with genetically different cell layers. Plant genomes vary widely in size, from less than 100 Mbp in some carnivorous plants such as Genlisea, to more than 17,000 Mbp in wheat and pines and on upwards, compared to human with 3,000 Mbp. Bananas are at the lower end of the range, with about 550 Mbp in the haploid set, and each chromosome is about 50 Mbp in size, the size of the human 21 or 22 chromosome. Figure 2 shows the chromosomes of ‘Cavendish’ labelled in situ with the 45S and 5S rDNA genes; each genome has one major site of the 45S tandemly repeated gene. Plant genomes, like those of animals, are full of repetitive DNA sequences and these vary in their sequence between the different genomes. Total genomic DNA from the wild species can be labelled and used as a probe to chromosomes of the cultivars to show the ancestral origin of these genomes. In figure 3, a metaphase of the ABB plantain ‘Bloggoe’ shows the 11 chromosomes from the A genome progenitor with the centromeric regions labelled in red, while the remaining 22 chromosomes come from the B genome. |
 |
|
| The mammalian genome is devoid of copia and gypsy-like retroelements that make up a substantial part of plant genomes, but in contrast, retroviruses and their recognizable remnants make up some 6% of the human genome but no DNA viruses with nuclear integration were known in plants until the late 1990s. At that time, there were several reports of a pararetrovirus (a group including hepatitis B) called Banana Streak Virus (BSV) appearing in plantations with no prior history of the disease, particularly where the plants had been stressed by either propagation through tissue culture or growing where night temperatures dropped below 22 ºC. PCR labs were able to identify presence of the virus, but were also amplifying fragments from control plants. Conventional explanations included the presence of virus particles in asymptomatic infected plants and PCR contamination. Fortunately, in situ hybridization of the virus sequence to root tip preparations could show that the virus was indeed located along the chromosomes in the nucleus (Figure 4). Furthermore, in situ hybrid-ization to extended DNA fibres made from the nucleus could show that the virus was integrated into the genome at a small number of sites with multiple copies in a complex arrangement, work that assisted the molecular cloning of the endogenous para-retroviral (EPRV) sequences. Now EPRVs are being found widely in plant genomes. |
|
Figure 1. a) A banana plant of the dessert variety ‘Cavendish’. Bananas are giant herbs and may be up to 5m tall, with the morphological structure of a grass, not a tree. A fruit bunch is harvested every 9 months or so, when the rest of the pseudostem is cut down and burned to control disease, being replaced by a sucker such as that seen at the base of the stem. b) Various triploid hybrid banana and plantain varieties on sale in India: from left to right: variety ‘Nendran’ (genome constitution AAB) used for cooking and for making chips, ‘Red’ (AAA), a prized sweet dessert banana cultivar, ‘Robusta’ (AAA, a ‘Cavendish’ type that is exported to the west; it becomes green-ripe at high temperatures,), ‘Nendran’ (AAB, ripe yellow, size difference from the first bunch is not genetic but due to water and fertilization differences), and ‘Peyan’ (ABB, rear) used as a vegetable for curries and for cooked snacks. c) Sweet and savoury banana chips and banana-based curries on a banana leaf plate. Figure 2-4. Metaphase chromosomes of triploid (2n=3x=33) banana cultivars stained blue with DAPI. Fig. 2) A metaphase of the export variety ‘Cavendish’ with AAA genome constitution, labelled by in situ hybridization with the 5S (green, b) and 45S rRNA (red, c) gene probes, showing one site of the 45S rDNA in each of the three genomes present in a triploid. Fig. 3) Metaphase chromosomes of the ABB plantain ‘Bloggoe’ shows the 11 chromosomes from the A genome progenitor with the centromeric regions labelled in red (b) by in situ hybridization with total genomic DNA from the wild Musa balbisiana (2n=2x=22), while the remaining 22 chromosomes come from the B genome (diagram in c). Figure 4. Metaphase chromosomes of ‘Obino L’Ewai’ (AAB) showing two chromosome pairs (arrowed) with sites of integration of the pararetrovirus Banana Streak Virus (red; b). |
How do the triploid plants arise? There are two mechanisms, most simply from a tetraploid crossing with a diploid. Tetraploids occur spontaneously and may be healthy and productive – many domesticated plants are selected as polyploids because they have larger fruit and more showy flowers. An alternative, more frequent mechanism to generate triploids is through production of 2n gametes. One of several stages of meiosis fails, giving what used to be called an ‘unreduced gamete’ containing a 2n chromosome complement from the parent through missing segregational or reductional divisions, formation of a restitution nucleus, or through a mitotic division replacing meiosis. Depending on how meiosis fails, the chromosome set may be identical to the parent, show segregation or include recombinant chromosomes. The gametes in the 2n pollen grains or 2n eggs are functional, and on fertilizing a normal, n, gamete they give triploid progeny.
The triploid bananas are sterile, and most sterile plants produce no fruits. However, the fruit in banana is produced without any seed, the mammalian equivalent of a placenta without fertilization occurring and without any embryo. This fruit development, known as parthenocarpy, involves a number of changes in hormonal control and its triggering by the stage of the fruit meristems rather than fertilization and zygote or endosperm development, although in banana the genetic mutations involved are not known. Interestingly, some banana cultivars have a very low rate of residual fertility, and a few seeds may be found by crossing hundreds or thousands of fertile diploids with sterile triploids. The fertility is probably the result of a few gametes having complete genomes through balanced genome segregation.
As with all agricultural crops, banana farmers face a continuous battle with newly evolving and spreading diseases. In the 1950s and 1960s, the then widespread export variety, ‘Gros Michel’, was devastated by Panama Disease, a fungal Fusarium wilt. Now, a variant known as Tropical Race 4 has become highly virulent on ‘Cavendish’; about 30% of production costs are for chemical controls, and 25 applications are typically required for one nine to 12 month production cycle. Fortunately there is extensive genetic variation within both wild A genome species and within the cultivars, and molecular marker methods are proving invaluable to measure this variation as a prelude to its exploitation. Using this variability is difficult with the sterility and lack of seeds, and because of widespread translocations suggested by preliminary cytogenetic work. 2n gametes, 4x X 2x hybridization, working with hundreds of fingers to find one seed, and transgenic strategies with well-characterized genes from heterologous species or potentially other banana varieties are now being used for breeding new cultivars. Molecular cytogenetics and the knowledge and understanding it gives is proving valuable in identifying parents, screening hybrids and tar-geting approaches for ensuring the sustainability and efficiency of banana as a crop for both smallholder and commercial farmers, with charac-ters including better disease resistance, quality, water use and yield.
Further details about banana can be found in the review Heslop-Harrison JS and Schwarzacher T. 2007. Domestication, genomics and the future for banana. Annals of Botany (2007) (Link to Heslop-Harrison JS, Schwarzacher T. 2007. Domestication, genomics and the future for banana). Our laboratory website, http://www.molecularcytogenetics.com/ gives additional results and references to original papers.
Address of the author: Pat Heslop-Harrison Department of Biology University of Leicester LEICESTER LE1 7RH, UK Tel.: +44 116 252 5079 Fax.: +44 116 252 2791 E-mail: phh4(a) le . ac . uk