|
Molecular cytogenetics website homepage -- Our Drosophila abstracts -- Our Drosophila papers -- Drosophila links including phylogeny Updates Sept 2011
Tandemly Repeated and Satellite DNA in the Drosophila buzzatii cluster
Prof. Dr. Gustavo Kuhn
Universidade Federal de Minas Gerais
Departamento de Biologia Geral (L3 164) Belo Horizonte, MG BRASIL
Tel: +55 31 3409-2599
Home page: www.icb.ufmg.br/gustavokuhn
e-mail: guto.cs.kuhn(a)googlemail.com or gcskuhn(a)ufmg.br
Guto Kuhn publications in PUBMED. Guto Kuhn publications/citations in Google Scholar.
Guto Kuhn's projects are described on his website at Universidade Federal de Minas Gerais
www.icb.ufmg.br/gustavokuhn . Here, we describe some project involving collaboration with Pat Heslop-Harrison and Trude Schwarzacher (University of Leicester, UK), Fabio M. Sene (Universidade de São Paulo, Brazil) and Orlando Moreira-Filho (Universidade Federal de São Carlos, Brazil) and others. |
|

Dr Gustavo Kuhn
Personal webpage: www.myspace.com/gustavokuhn |
The genome of eukaryotic organisms is replete with a variety of repetitive DNA elements that do not encode products used by the cell. They are classified into two major classes according to their genomic organization. The first class is composed of DNA motifs of various lengths and composition that occur as tandem repeats that are localized at one or more distinct positions in the genome. The second class is represented by sequences with a dispersed distribution throughout the genome. The majority of these “tandem” or “dispersed” repeats are represented by satellite DNAs and transposable elements.
Despite extensive study in several organisms, there are still many questions remaining unsolved, especially concerning satDNA evolution, function and contribution to speciation.
Our research has focused on the following areas:
-
Patterns and mechanisms of satellite DNA evolution (effects of DNA sequence, abundance, chromosome location, array sizes and long-range organization)
-
The contribution of transposable elements to the evolution of genomes in Drosophila species
-
The large-scale evolution of Drosophila genomes and individual chromosomes
Model species include the cactus-breeding Drosophila species from the largely South American Drosophila buzzatii cluster (repleta group) and the 12 sequenced Drosophila genomes. This programme addresses important areas of genome evolution, speciation and phylogenetics in relation to mechanisms of sequence turnover and species differentiation in the natural environment. |
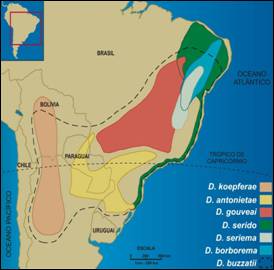
The distribution of the seven species from the Drosophila buzzatii cluster in South America Species from the buzzatii cluster are cactus-breeding and occur on natural environments. |
 |
| Drosophila seriema (buzzatii cluster, repleta group) DNA fibres hybridised with two satellite DNA probes, alpha/beta (green signals) and DBC-150 (red signals). The picture shows several long satellite DNA arrays pulling out of the nucleus (right) |
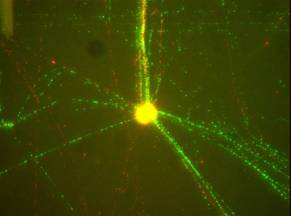
|
We have been studying different satellite DNAs in a group of seven closely related cactus-breeding Drosophila species from South America, the buzzatii cluster (repleta group). These species are special because they share a number of satellite DNAs showing different characteristics, such as repeat length, DNA structure, chromosomal distribution, array sizes and organization, abundance, variability and homogenization rates. We are investigating five satellite DNAs, pBuM-1 (monomer length 190 bp), pBuM-2 (370 bp), DBC-150 (150 bp) and two additional new ones (300 bp and 400 bp).
|
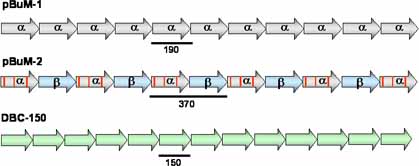 |
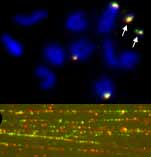
|
In situ hybridization shows on chromosomes (upper panel) that two tandemly repeated sequences are present on some of the chromosomes: the microchromosomes, the X and one pair of autosomes. The lower panel shows in situ hybridization of the same sequences to DNA fibres with both interspesed and single tandem arrays of the two sequences.
This is a multi-disciplinary project that was build to give insights towards a better understanding of how satellite DNAs evolve and contribute for species genomic cohesion and evolution. We are also initiating projects to analyse satellite DNAs in the genomes of the 12 sequenced Drosophila genomes. |
Publications - Link to Drosophila publication page: (Most title links need intranet password - DOI links need subscription)
279. Kuhn GC, Schwarzacher T, Heslop-Harrison JS. 2010. The non-regular orbit: three satellite DNAs in Drosophila martensis (buzzatii complex, repleta group) followed three different evolutionary pathways. Molecular Genetics and Genomics 284(4): 251-262. http://dx.doi.org/10.1007/s00438-010-0564-1.
268. Kuhn GCS, Teo CH, Schwarzacher T, Heslop-Harrison JS. 2009. Evolutionary dynamics and sites of illegitimate recombination revealed in the interspersion and sequence junctions of two nonhomologous satellite DNAs in cactophilic Drosophila species. Heredity 102: 453–464. doi: 10.1038/hdy.2009.9 (The link to the title contains the full colour plate - also available here - which is not in the Heredity DOI for the PDF but is in the HTML version; it is rather essential to show the results in the paper.)
255. Kuhn GCS, Sene FM, Moreira-Filho O, Schwarzacher T, Heslop-Harrison JS. 2008. Sequence analysis, chromosomal distribution and long-range organization show that rapid turnover of new and old pBuM satellite DNA repeats leads to different patterns of variation in seven species of the Drosophila buzzatii cluster. Chromosome Research 16: 307-324. DOI link: 10.1007/s10577-007-1195-1
Kuhn GCS, Franco, FF, Manfrin MH, Moreira-Filho O & Sene FM (2007). Low rates of homogenization of the DBC-150 satellite DNA family restricted to a single pair of microchromosomes in species from the Drosophila buzzatii cluster. Chromosome Res. 15(4): 457-470.
Franco FF, Kuhn GCS, Sene FM & Manfrin MH (2006). Conservation of pBuM-2 satellite DNA sequences in isolated populations of Drosophila gouveai from Brazil. Genetica 128(1-3): 287-95.
Kuhn GCS & Sene FM (2005). Evolutionary turnover of two pBuM satellite DNA subfamilies in the Drosophila buzzatii cluster (repleta group): from alpha to alpha/beta arrays. Gene 349: 77-85.
Kuhn GCS & Sene FM (2004) Characterisation and Interpopulation Variability of a complex HpaI Satellite DNA of Drosophila seriema (repleta group). Genetica 121(3): 241-249.
Kuhn GCS, Franco FF, Silva Jr WA, Rossi NMM & Sene FM (2003). On the pBuM189 satDNA variability in D. buzzatii populations in South-America. Hereditas 139: 161-166.
Kuhn GCS, Bollgönn S, Sperlich D, Bachmann L (1999). Characterization of a species-specific satellite DNA of Drosophila buzzatii. J. Zool. Syst. Evol. Res. 37: 109-112. |
Please send corrections, comments
or questions to guto.cs.kuhn(at)googlemail.com
|