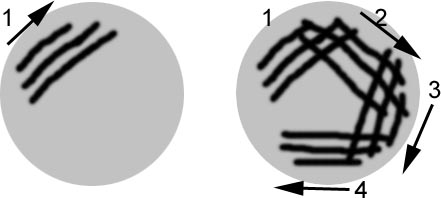
3. Place plates at 37C overnight to grow.
4. Ideal appearance is continuous line of colonies in lines 1, going to individual colonies clearly separated in lines 4
5. Pick individual colonies for isolation of BACs by pushing a sterile toothpick into the colony.
Small-scale isolation of BAC DNA from 10 ml cultures
Based on protocol from Zhang from Texas A&M University - http://hbz.tamu.edu/manual.pdf
Particular points to note are gentle shaking and inversion during the DNA isolation: these lead to differential precipitation of unwanted bacterial genomic DNA (4.6 MegaBases) and the BAC DNA (40 to 200 kiloBases) and keep the DNA in large molecules.
1. Stab a single colony from the agar plate above with a sterile toothpick and drop into 10 ml LB medium (or 20 ml, Abbott) with 25 ul chloramphenicol solution (50 mg/ml) in a 50ml tube. (This is 10X higher than ofen used in Abbott's lab.)
(A fast and dirty method is to use 10ul of a liquid culture for the inoculation but this is not recommended as it is important that a colony from a single cell is grown up to ensure purity)
2. Grow overnight with rapid shaking at 37C - 24 hr is good for the culture period.
3. Centrifuge culture at 1500g for 10 min, discard supernatant into waste container containing a few ml of bleach, and resuspend the pellet in the remaining 100 ul of culture medium. Add 100 ul of lysis buffer (50 mM glucose, 10 mM EDTA pH 8.0, 25 mM Tris.HCl pH8, autoclaved and stored at 4C; Abbott lab: Add 4.5 ul of 10mg/ml RNase), shake gently and incubate on ice for 5 min. (Abbott lab: remove all culture medium that is possible and add 300 ul of GTE/RNase, resuspend by hard vortexing.)
4. Add 400 ul of 0.2 M NaOH, 1% SDS solution, shake gently and put on ice for 5 min (Abbott lab: 300 ul for a few seconds: as long as 5 min will denature BAC DNA irreversibly.).
5. Add 300 ul of 3M KOAc (potassium acetate adjusted to pH 5 with acetic acid) and incubate on ice for 15 min. (Abbott: not necessary to wait as long.)
6. Centrifuge at 2800g for 15 min. (Abbott: transfer to 2ml microcentrifuge tubes and spin 15000 g 15 min.)
7. Transfer supernatant (c. 750 ul) to microcentrifuge tube, add 450 ul isopropanol (Abbott: equal volume of isopropanol), mix and centrifuge at 12000g for 5 min.
8. Discard supernatant, wash pellet with 70% ethanol, centrifuge 12000g for 2 min.
9. Invert tube over a tissue and tip out ethanol and allow pellet to air dry for 20 min (less in dry atmosphere - it should not become fully dry)..
10. Resuspend pellet in 40 ul TE (pH 8) or water, allowing to redissolve at room temperature overnight or by heating up to 65C. Do not vortex! (Resuspending in water means the DNA only lasts a few weeks at -20C; using TE inhibits future restriction digestions; Abbott lab: suggests 0.1 mM Tris pH 8 for resuspension.)
11. Treat with RNase:: add 2ul of 10mg/ml DNase-free RNase and incubate for 30 min at 37C.
12. Make a test digest of the BAC DNA:
10 ul of BAC DNA solution in TE/water
25.5 ul water (molecular biology grade, purchased from Sigma etc)
4 ul 10x restriction enzyme buffer
0.5 ul of restriction enzyme ( Hae III is often a good choice)
Digest overnight (min 3 hours) at 37C and run 10 ul in loading buffer on 0.8% agarose gel (0.5% can be better as it will separate fragments .5kb better), stain with ethidium bromide and photograph. Each lane of BAC DNA should have a number of discrete bands. It should digest almost completely (no DNA remaining in well), there should be no RNA (diffuse, large smear at bottom of gel) and each BAC should have a different and characteristic 'fingerprint' (pattern of bands).
Thanks to Sam Forrest from Bert Abbott's lab, South Carolina for comments on the protocol.
Pat Heslop-Harrison
University of Leicester LE1 7RH UK
www.molcyt.com
phh4 @ REMOVE le.ac.uk |