![]() QLTR 2001-02098
QLTR 2001-02098
Pararetroviruses, diseases, integration and genomes

October 2002
until April 2006
PARADIGM
is a programme in the EU framework V
coordinated by Marie-Line Caruana and
Pierre-Yves Teycheney of the Centre International de Recherche Agronomique pour le Développement (CIRAD), France, and involves seven partners
from France, UK, Austria, Switzerland and Spain.
Project
duration: October 2002 – April 2006
Numerous
endogenous pararetroviruses (EPRVs) are found to be
integrated in variable copy numbers into the genome of various crops such as
banana, petunia and tobacco. Although EPRVs might in some cases be simply
neutral components of plant genomes, in other instances they can be infectious,
express viral particles and cause severe diseases in plants. Recent
experimental evidences show that EPRVs are widespread in the genome of a large
number of crops and might be highly detrimental to existing and future crop
improvement-oriented breeding programmes, in vitro mass propagation of
vegetative crops and germplasm conservation. Our
project aims at studying the biodiversity and roles of EPRVs in plant germplasms, their interactions with plant and viral
genomes, the associated risks in pathogenesis, and their potential to confer
viral resistance in order to develop appropriate risk assessment strategies for
the control of potentially pathogenic EPRV sequences in crops relevant to
European agriculture.
Overall objectives of the PARADIGM consortium
The overall research that is carried out by the consortium will be primarily based on existing experimental models (banana, tobacco, petunia) for the development of risk assessment strategies, then applied to crops relevant to European agriculture for the implementation stage. The consortium will share and apply their acknowledged complementary expertise and experience to develop experimental approaches with the specific objectives of:
· Investigating the biodiversity and evolutionary status of EPRVs in such crops;
· Screening economically important crops to European agriculture for possibly deleterious EPRVs;
· Evaluating the risks of EPRVs for germplasm conservation and for ongoing and future breeding programmes, including those aimed at creating genetically modified crops;
· Determining the mechanisms of (i) integration of EPRVs in plant genomes and (ii) those of activation, excision and expression of EPRVs leading to infectious episomal viral genomes;
· Defining the effect of EPRVs on plant genomes stability and dynamics;
· Characterizing the mechanisms developed by plants to control EPRVs, such as gene silencing, and the strategies developed by pararetroviruses to suppress such controls;
·
Devising risk assessment and control strategies
for EPRVs, and implementing such strategies through industrial partnerships
with European plant biotech and plant breeding
![]()
To address these concerns, the PARADIGM project has been divided into 4 scientific workpackages aimed at answering clearly defined questions:
WPI: What
is the exact extent and biodiversity of EPRVs within plant germplasms?
WPII: What are the mechanisms of movement of viral sequences into and out of the plant genomes?
WPIII: What are the interactions between EPRVs and
plant genomes?
WPIV: What is the role of, and risk related to, EPRVs
in pathogenesis?
![]()
The team in
Leicester, lead by Dr Trude Schwarzacher,
was coordinating WPI and plays a
leading role in examining the organisation and diversity of EPRV and related
sequences in the host plant nuclear genome. Together with other partners, we
are developing universal tools to isolate EPRVs from a limited number of crops
where EPRV activation has already been observed, and investigate the
biodiversity represented in EPRV sequences. By examination of short and long
clones, and by PCR, we plan to determine the nature, organisation and sequence
relationships of EPRVs between accessions of two target species groups,
examining the copy numbers and chromosomal arrangement, long-range organisation
and fine structure of EPRVs. Partners will develop evolutionary and structural
models of EPRVs that will allow us to predict infection and expression routes.
In the final task, partners will design molecular tools for identifying
candidate EPRVs in any species, concentrating on five major European crops and,
attempt implement them for routine screening; results, probes and primers will
be delivered. We will develop S-SAP-based and inverse PCR (I-PCR) approaches
for the characterisation of EPRVs integration sites and develop methods for the
analysis of longer range flanking sequences.
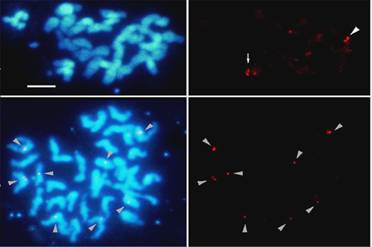

Banana chromosomes stained with DAPI (blue) and
after in situ hybridization with Banana Streak Virus probe (red). Two to
eight integration sites have been detected (arrows). See Harper et al. (1999)
for details.
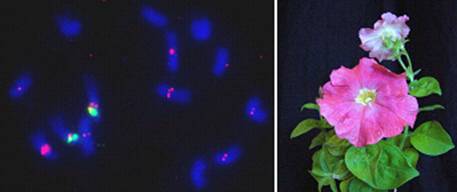
Chromosomes of Petunia hybrida ‘Himmelsroeschen’ after
DAPI staining (blue) and in situ hybridization with Petunia Vein
Clearing Virus probe (red) and 5Sr DNA (green). See Richert-Poeggeler
et al. (2003) for more detail.
Recent publications relevant to the subject:
Harper, G., Osuji, J.O.,
Heslop-Harrison, P. Hull, R. (1999).
Integration of banana streak badnavirus into the Musa genome : molecular evidence. Virology 255: 207-213.
Teo
CH, Tan SH, Othman YR, Schwarzacher T (2002) The
cloning of Ty 1-copia-like retrotransposons from 10 varieties of banana (Musa
sp). Journal of Biochemistry, Molecular Biology and Biophysics 6,193-201.
Richert-Pöggeler, K.R., Noreen, F., Schwarzacher,
T., Harper, G. and Hohn, T. (2003) Induction of
infectious Petunia vein clearing (pararetro) virus
from endogenous provirus in petunia. EMBO Journal 22: 4836-4845. doi: 10.1093/emboj/cdg443
Hansen CN, and Heslop-Harrison JS (2004) Sequences and
Phylogenies of Plant Pararetroviruses, Viruses and
Transposable Elements. Advances in
Botanical Research ISBN-13: 978-0-12-005941-6 41:
165-193
Hansen CN, Harper G, and Heslop-Harrison JS (2004)
Characterization of pararetrovirus-like sequences in the genome of potato (Solanum tuberosum). Cytogenet Genome
Res 559-565. DOI:
10.1159/000084989
Staginnus,
C., Gregor, F., Mette, M.F.,
Hohn T, Richert-Pöggeler
KR, Staginnus C, Harper G, Schwarzacher T, Chee How
Teo CH, Teycheney P-Y, Iskra-Caruana
ML, Hull R (2008). Evolution of integrated plant viruses.
In: Plant virus evolution. (Rossinck
M, ed.), Springer, Berlin, ISBN 978-3-540-75762-7. pp 53-82.
Schwarzacher T
(2008) Fluorescent In Situ Hybridization to Detect Transgene Integration into Plant Genomes. in Transgenic Wheat, Barley and Oats. Huw
D. Jones and Peter R. Shewry (eds.), Methods in
Molecular Biology, 478,
227-246 doi: 10.1007/978-1-59745-379-0